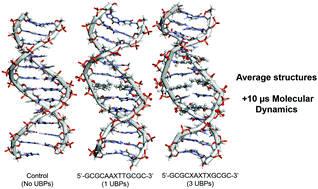
In vivo inclusion of unnatural base pairs (UBPs) into functional DNA was recently reported for compounds 2,6-dimethyl-2H-isoquiniline-1-thione (d5SICS, X) and 2-methoxy-3-methylnaphthalene (dNaM, Y) in a modified E. coli strand, for which high-fidelity replication was observed. Yet, little is known about possible genetic consequences they have in largely substituted DNA. Using a converged microsecond long molecular dynamics (MD) simulation of the sequences 5′-GCGCAAXTTGCGC-3′ and 5′-GCGCXAXTXGCGC-3′, where X represents the UBP, we show that in the system with only a single XY UBP pair present, the global RMS deviation from canonical B-DNA in our control simulations is ∼3 Å and a fully converged ensemble is achieved within 2 µs. With three UBPs, deviation increases to ∼5 Å and convergence is not achieved within 10 µs of sampling time. With five UBPs, no convergence is observed and the double helix collapses into a globular structure. A fully optimized structure of the trimer d(GXC) was obtained using the density functional theory method B97D/cc-pVTZ and resulted in an RMSD value of ∼2 Å when compared to the most populated structure obtained from the MD simulations. Their calculated interaction energy is −3.7 kcal mol−1. It is thus unlikely that d5SICS and dNaM could be useful as tools in DNA manipulation. This theoretical methodology can be used in the de novo design of UBPs.